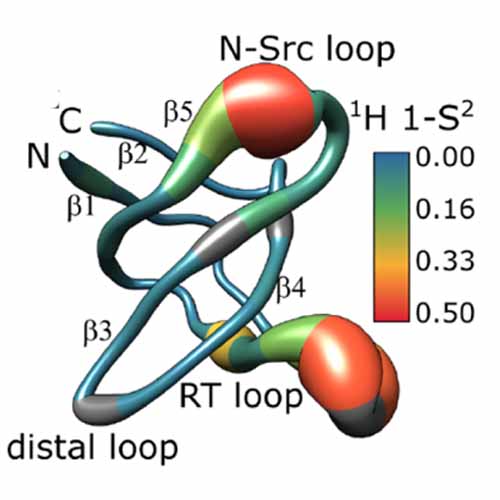Mechanistic Insights into Microsecond Time-Scale Motion of Solid Proteins Using Complementary 15N and 1H Relaxation Dispersion Techniques
2019-01-08
Petra Rovó, Colin A. Smith, Diego Gauto, Bert L. de Groot, Paul Schanda, and Rasmus Linser
J. Am. Chem. Soc., 2019, 141, 2, 858–869
NMR relaxation dispersion methods provide a holistic way to observe microsecond time-scale protein backbone motion both in solution and in the solid state. Different nuclei (1H and 15N) and different relaxation dispersion techniques (Bloch–McConnell and near-rotary-resonance) give complementary information about the amplitudes and time scales of the conformational dynamics and provide comprehensive insights into the mechanistic details of the structural rearrangements. In this paper, we exemplify the benefits of the combination of various solution- and solid-state relaxation dispersion methods on a microcrystalline protein (α-spectrin SH3 domain), for which we are able to identify and model the functionally relevant conformational rearrangements around the ligand recognition loop occurring on multiple microsecond time scales. The observed loop motions suggest that the SH3 domain exists in a binding-competent conformation in dynamic equilibrium with a sterically impaired ground-state conformation both in solution and in crystalline form. This inherent plasticity between the interconverting macrostates is compatible with a conformational-preselection model and provides new insights into the recognition mechanisms of SH3 domains.