Strategies to Avoid Artifacts in Mass Spectrometry-Based Epitranscriptome Analyses
2021-08-02
Dr. Steffen Kaiser, Dr. Shane R. Byrne, M. Sc. Gregor Ammann, Paria Asadi Atoi, Dr. Kayla Borland, Dr. Roland Brecheisen, Dr. Michael S. DeMott, Dr. Tim Gehrke, M. Sc. Felix Hagelskamp, M. Sc. Matthias Heiss, M. Sc. Yasemin Yoluç, Lili Liu, Qinghua Zhang, Prof. Peter C. Dedon, Prof. Bo Cao, Prof. Dr. Stefanie Kellner
Angew. Chem. Int. Ed., 2021, Volume 60, Issue 44, Pages 23885–23893
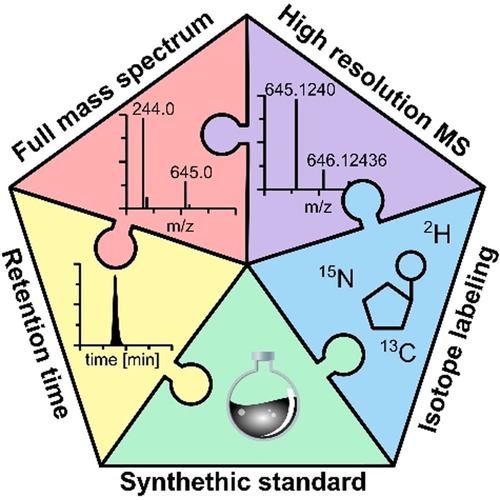
In this report, we perform structure validation of recently reported RNA phosphorothioate (PT) modifications, a new set of epitranscriptome marks found in bacteria and eukaryotes including humans. By comparing synthetic PT-containing diribonucleotides with native species in RNA hydrolysates by high-resolution mass spectrometry (MS), metabolic stable isotope labeling, and PT-specific iodine-desulfurization, we disprove the existence of PTs in RNA from E. coli, S. cerevisiae, human cell lines, and mouse brain. Furthermore, we discuss how an MS artifact led to the initial misidentification of 2′-O-methylated diribonucleotides as RNA phosphorothioates. To aid structure validation of new nucleic acid modifications, we present a detailed guideline for MS analysis of RNA hydrolysates, emphasizing how the chosen RNA hydrolysis protocol can be a decisive factor in discovering and quantifying RNA modifications in biological samples.