The Enzymatic Decarboxylation Mechanism of 5-Carboxy Uracil: A Comprehensive Quantum Chemical Study
2020-12-24
Andrea Kreppel and Christian Ochsenfeld
J. Chem. Theory Comput. 2021, 17, 1, 96 – 104
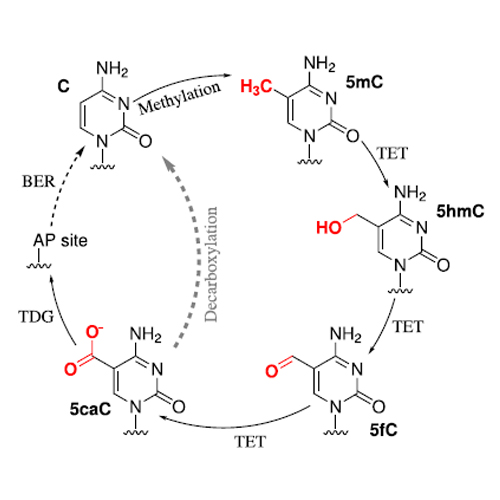
Dynamic regulation of DNA methylation is an important process for the control of gene expression in mammals. It is believed that in the demethylation pathway of 5-methyl cytosine, the intermediate 5-carboxy cytosine (5caC) can be actively decarboxylated alongside the substitution in the base excision repair. For the active decarboxylation of 5caC, a decarboxylase has not been identified so far. Due to the similar chemistry of the decarboxylation of 5-carboxy uracil (5caU) to uracil (U) in the pyrimidine salvage pathway catalyzed by the iso-orotate decarboxylase (IDCase), the study of this reaction might give valuable insights into the active 5caC decarboxylation process. In this work, we employ quantum chemical and molecular mechanic calculations and find that the catalytic mechanism of IDCase proceeds via a direct decarboxylation mechanism. Detailed investigations on the reaction coordinate reveal that it is a one-step mechanism with concerted proton transfer and C–C bond opening.