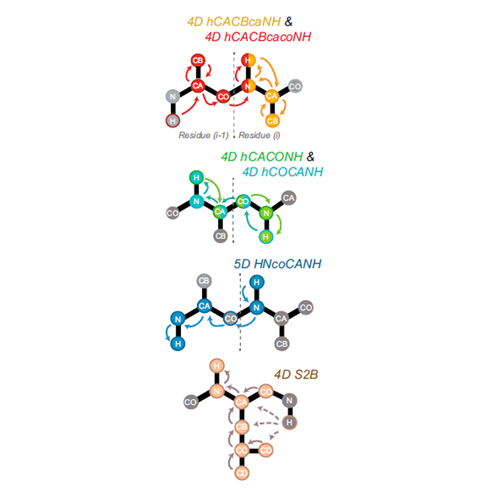
Atomic-resolution chemical characterization of (2x)72-kDa tryptophan synthase via four- and five-dimensional 1H-detected solid-state NMR
2022-01-20
Alexander Klein, Petra Rovó, Varun V. Sakhrani, Yangyang Wang, Jacob B. Holmes, Viktoriia Liu, Patricia Skowronek, Laura Kukuk, Suresh K. Vasa, Peter Güntert, Leonard J. Mueller, Rasmus Linser
NMR chemical shifts provide detailed information on the chemical properties of molecules, thereby complementing structural data from techniques like X-ray crystallography and electron microscopy. Detailed analysis of protein NMR data, however, often hinges on comprehensive, site-specific assignment of backbone resonances, which becomes a bottleneck for molecular weights beyond 40 to 45 kDa. Here, we show that assignments for the (2x)72-kDa protein tryptophan synthase (665 amino acids per asymmetric unit) can be achieved via higher-dimensional, proton-detected, solid-state NMR using a single, 1-mg, uniformly labeled, microcrystalline sample. This framework grants access to atom-specific characterization of chemical properties and relaxation for the backbone and side chains, including those residues important for the catalytic turnover. Combined with first-principles calculations, the chemical shifts in the β-subunit active site suggest a connection between active-site chemistry, the electrostatic environment, and catalytically important dynamics of the portal to the β-subunit from solution.