Decrypting drug actions and protein modifications by dose- and time-resolved proteomics
2023-03-16
Jana Zecha, Florian P. Bayer, Svenja Wiechmann, Julia Woortman, Nicola Berner Julian Müller, Annika Schneider, Karl Kramer, Mar Abril-Gil, Thomas Hopf, Leonie Reichart, Lin Chen, Fynn M. Hansen, Severin Lechner, Patroklos Samaras, Stephan Eckert, Ludwig Lautenbacher, Maria Reinecke, Firas Hamood1, Polina Prokofeva, Larsen Vornholz, Chiara Falcomatà, Madeleine Dorsch, Ayla Schröder, Anton Venhuizen, Stephanie Wilhelm, Guillaume Médard, Gabriele Stoehr, Jürgen Ruland, Barbara M. Grüner, Dieter Saur, Maike Buchner, Benjamin Ruprecht, Hannes Hahne, Matthew The, Mathias Wilhelm, Bernhard Kuster
J. Zecha et al., Science, 2023
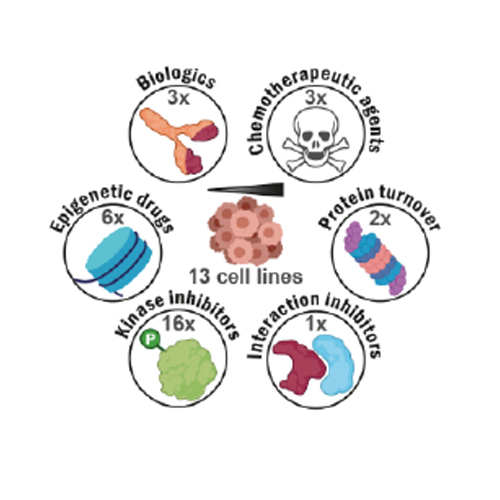
Although most cancer drugs modulate the activities of cellular pathways by changing post-translational modifications (PTMs), surprisingly little is known regarding the extent and the time- and dose-response characteristics of drug-regulated PTMs. Here, we introduce a proteomic assay termed decryptM that quantifies drug-PTM modulation for thousands of PTMs in cells to shed light on target engagement and drug mechanism of action (MoA). Examples range from detecting DNA damage by chemotherapeutics, to identifying drug-specific PTM signatures of kinase inhibitors, to demonstrating that rituximab kills CD20-positive B-cells by over-activating B cell receptor signaling. DecryptM profiling of 31 cancer drugs in 13 cell lines demonstrates the broad applicability of the approach. The resulting 1.8 million dose-response curves are provided as an interactive molecular resource in ProteomicsDB.