The type of DNA damage response after decitabine treatment depends on the level of DNMT activity
2024-06-21
Tina Aumer, Maike Däther, Linda Bergmayr, Stephanie Kartika, Theodor Zeng, Qingyi Ge, Grazia Giorgio, Alexander J Hess, Stylianos Michalakis, Franziska R Traube
Life Science Alliance, vol.7, no9, e202302437, 2024
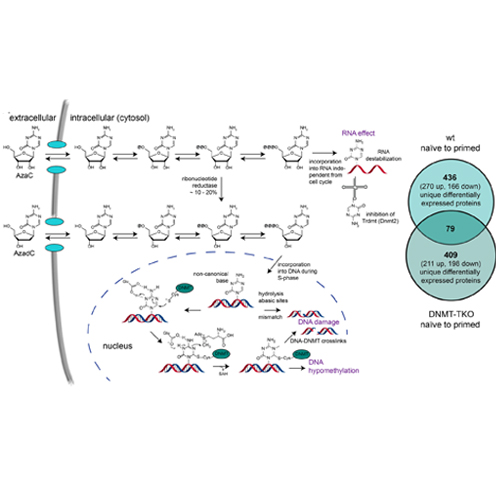
Decitabine and azacytidine are considered as epigenetic drugs that induce DNA methyltransferase (DNMT)-DNA crosslinks, resulting in DNA hypomethylation and damage. Although they are already applied against myeloid cancers, important aspects of their mode of action remain unknown, highly limiting their clinical potential. Using a combinatorial approach, we reveal that the efficacy profile of both compounds primarily depends on the level of induced DNA damage. Under low DNMT activity, only decitabine has a substantial impact. Conversely, when DNMT activity is high, toxicity and cellular response to both compounds are dramatically increased, but do not primarily depend on DNA hypomethylation or RNA-associated processes. By investigating proteome dynamics on chromatin, we show that decitabine induces a strictly DNMT-dependent multifaceted DNA damage response based on chromatin recruitment, but not expression-level changes of repair-associated proteins. The choice of DNA repair pathway hereby depends on the severity of decitabine-induced DNA lesions. Although under moderate DNMT activity, mismatch (MMR), base excision (BER), and Fanconi anaemia–dependent DNA repair combined with homologous recombination are activated in response to decitabine, high DNMT activity and therefore immense replication stress induce activation of MMR and BER followed by non-homologous end joining.